Exploring Spatial Biology With Dr. Nigel Jamieson
Spatial biology allows the visualization of cells and molecules within their unique two- or three-dimensional contexts and could revolutionize our understanding of health and disease.
Complete the form below to unlock access to ALL audio articles.
Dr. Nigel Jamieson, clinical senior lecturer and honorary hepato-pancreato-biliary surgeon at Glasgow Royal Infirmary, is a pioneer in the field of spatial biology. He has made significant contributions to the understanding of spatial organizations and interactions within biological systems. His primary area of research focuses on the interactions of the genomic landscape, the immune system and chemotherapy in patients with pancreatic cancer.
Technology Networks invited Dr. Nigel Jamieson to an Ask Me Anything session to answer your questions about spatial biology. These are just some of the questions that we asked Nigel, click below to watch the full AMA.
Lucy Lawrence (LL): How does spatial biology impact our understanding of diseases like cancer, Alzheimer's or other health conditions?
Nigel Jamieson (NJ): For a long time, we've tried to understand diseases like cancer by looking at a lump of tissue. Within a square centimeter, there are many cells with complex relationships and a pattern of genes that are switched on or off. When you mash them all up and extract the RNA from them to try and discover what genes are expressed, the cellular anatomy and cellular geography are lost.
It’s important to understand not only what type of cells they are, but also their spatial orientation with respect to each other.
Until recently, we were limited to using complex technologies, such as transcriptomics, within frozen tissue or fresh tissue. This is incredibly difficult to do outside the controlled laboratory environment, with preclinical or patient tissue. Over the last couple of years, we’ve been able to analyze paraffin-embedded tissue – tissue preserved in wax blocks that pathologists use to make a diagnosis. The paraffin embedding process has made it much easier for us to analyze human samples using technologies like transcriptomics and spatial biology. This allows us to examine tissue architecture and contributes to our understanding of disease.
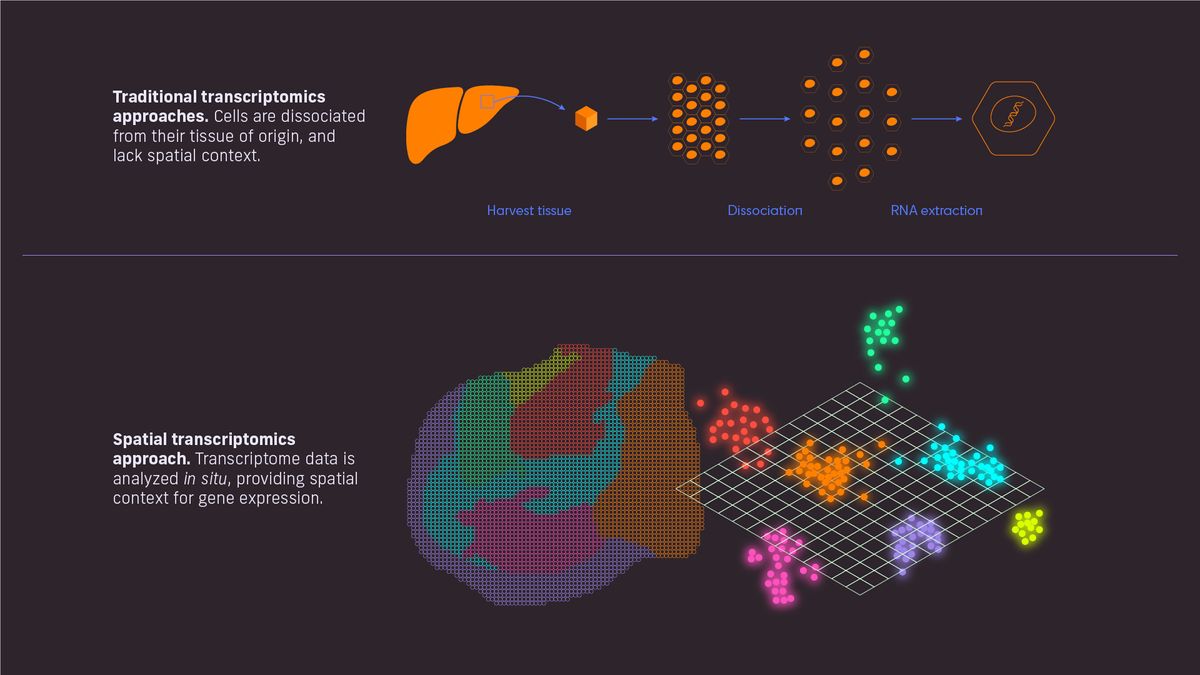
Figure 1: Traditional vs spatial transcriptomics. Credit: Technology Networks, adapted from Spatial Transcriptomics.
LL: Can spatial biology help us to understand the impact of lifestyle choices like diet and exercise on our overall health?
NJ: I think so, yes. In our group at the University of Glasgow and in collaboration with the Cancer Research Institute, we've been using spatial biology strategies to look at the endothelium of blood vessels and arteries.
By analyzing the cellular landscape and cellular interactions in tissue samples from human specimens, it is possible to study the impact of cardiovascular disease in an artery. You could also look at the impact of a fatty diet on the liver by studying a biopsy, for example. While we have been able to do that for a long time, it would have previously been harder to look at in a paraffin-embedded sample, and in large cohorts of samples.
Some of the new strategies enable us to study cohorts of tissue samples rather than just a few fresh samples. There’s now potential to get really detailed cellular interaction information and conduct wide transcriptomic analysis using a biopsy specimen. For example, spatial analysis of the kidneys has produced impressive data. You can see the glomeruli of the kidney overlaid with cells of interest and perform localized single-cell analysis within paraffin-embedded tissue.
I come at this from a cancer perspective, but ultimately, it's a huge opportunity for any sort of disease type to study both the normal anatomy and the perturbance of a disease.
LL: How does spatial biology intersect with immunology and oncology?
NJ: In oncology, traditional transcriptomics analyses are challenging because in a biopsy tissue, there are going to be areas that are mostly cancer cells, areas that are mostly normal cells and interfaces between the two. There are also a lot of immune cells present in both the normal and the cancerous areas of the biopsy. When you homogenize a sample to analyze it, the immune cells from each area of the tissue are mixed up, and it will be unclear where in the sample they were situated.
Additionally, immune cells coalesce to to educate each other about cancer cells through antigen presentation in immune tissue such as lymph nodes or tertiary lymphoid structures. Therefore, biopsies of immune tissue can suggest falsely high immune signaling and inaccurate results. Having the ability to not only see the signaling networks and where cells are coming from, but also their proximity to cancer cells, is extremely important.
In precision oncology, the expression of certain proteins on the surface of tumor cells influences the type of immunotherapies that cancer patients are given. We’ve been utilizing this for some time with lung cancers. Lung biopsies are taken and the expression of immunosuppressive proteins such as immune checkpoint markers are assessed. For example, if PD-L1 (which can switch off T cell activity) is present on the cancer cells, then the immune checkpoint inhibitor against PD-L1 can be administered. Spatial biology strategies can help with these approaches by giving closer insight into the protein expression on cancer cells and whether ligands, such as PD-L1, are interacting with their receptors on immune cells.
If you were to take a traditional approach to a biopsy and homogenize the sample, you would be able to tell if the ligand and receptor were present, but not if they were actually interacting. Spatial biology allows for far more accurate, and hopefully more successful, treatment strategies.
LL: Can spatial biology be applied in different fields, for instance, medicine, agriculture or environmental sciences?
NJ: Definitely. Ultimately, we can do human studies to help us understand human disease but at the same time, we can do animal studies to understand animal diseases.
I think it’s absolutely possible that spatial biology will transcend disease, into things like environmental processes, where we can see the implications of that on a cellular spatial level.
The concept of spatial biology is not new biology, it's just a way of potentially gaining more information and unlocking the geography of a process.
LL: Have you come up against any ethical considerations in your area of work?
NJ: A lot of the samples we work with are from preclinical experiments using mouse models. Therefore, we need to try and reduce the use of animals as much as possible and as ethically as possible. We really want to try and get as much information in the shortest amount of time using the least resources – both monetarily and in terms of using as little animal tissue as possible.
It’s also really important that we don’t waste tissue samples donated by patients for research. We can now study one tiny tissue sample and collect information on all the genes, or hundreds of proteins. In my opinion, this is much more ethical than the 100 sections we would have had to take previously to gather the same amount of data. We make sure we’re using the latest and best strategies to get as much information as possible. At the same time, better technology could come along in six months which could do the same analyses with half as much tissue. However, if all the tissue samples have already been used up, then you can’t utilize the latest and potentially more useful technology, so a balance does need to be found.
LL: How do you see the field of spatial biology evolving in the next few years?
NJ: I think the technologies will become easier to use, certainly. Costs are likely to come down, imaging resolution will increase, and the potential uses will broaden. I think a lot of the development will come down to normalizing the idea of spatial biology and making it more accessible to more researchers so that the techniques and strategies can replace lower-plex workflows.
Ultimately with spatial biology techniques, you’re generating more data for every sample analyzed, so costs will decrease, and it will work out cheaper to analyze the whole genome, or 5,000 genes, than to analyze 100 genes three times. I also think that the three-dimensional aspects of spatial biology are going to grow and improve, along with non-destructive tissue analysis. That means scanning a tissue sample in order to analyze it, so that information is gathered through imaging rather than destructively processing it, and the geography of the sample is better retained.
We may eventually be able to get all the cellular information that we’re currently gathering through spatial transcriptomics and proteomics just by scanning a biopsy sample using light field microscopy.
Dr. Nigel Jamieson was speaking to Lucy Lawrence, Senior Digital Content Producer for Technology Networks.
About the interviewee
Nigel Jamieson, Clinical Senior Lecturer at the Institute of Cancer Sciences and Honorary HPB surgeon at Glasgow Royal Infirmary, is a distinguished figure in pancreatic cancer research. Jamieson’s primary research interest focuses on the interplay between the genomic landscape, the immune microenvironment, and chemotherapy in patients diagnosed with pancreatic cancer. Specifically, he works to develop combination therapies that target immune evasion.