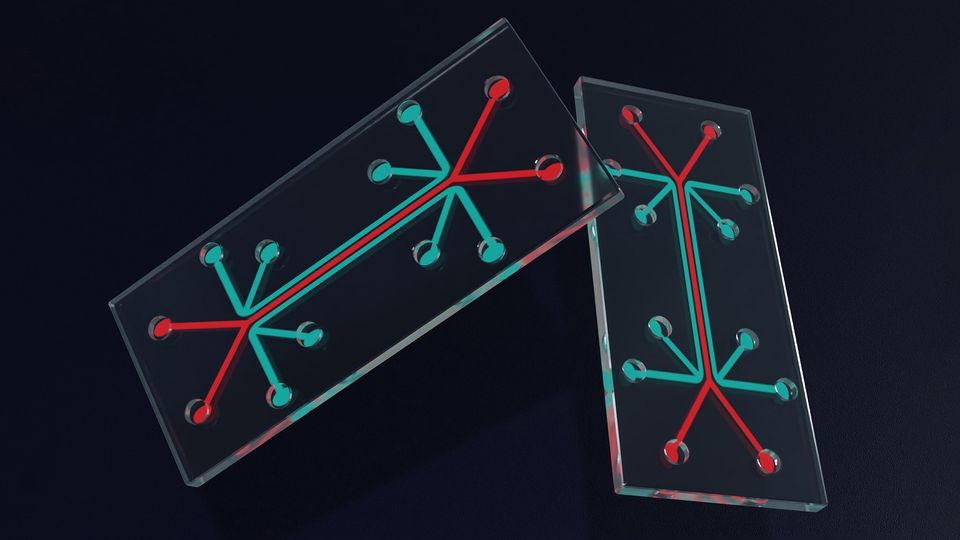
Advances in Organ-on-a-Chip Technology
Technological advances and changes in the regulatory landscape mean organ-on-a-chip systems could soon transform the way medicines are tested.
Complete the form below to unlock access to ALL audio articles.
An organ-on-a-chip (OOAC) is a model system containing engineered cells grown inside microfluidic chips.1 Although the concept of an OOAC is not new, technological advances and changes in the regulatory landscape mean they could soon transform the way medicines are tested.
Until recently, every single new medicine had to be tested preclinically in animal models, but now – in the US at least – the FDA Modernization Act 2.0 has paved the way for the use of alternatives, and OOACs are one option being explored for accelerating drug discovery and development.2
“OOACs offer the potential to solve clinical development attrition by providing human models that truly replicate the complex biological, biochemical and biomechanical environments you'd find in vivo, rather than being limited to what you can model in a petri dish,” says Professor Martin Knight, co-director of the Queen Mary Centre for Predictive in vitro Models and the UK Organ-on-a-Chip Technologies Network. “But there [are] key challenges to getting to that stage including how to represent the full complexity of human tissues and organs in both health and disease and identifying ways to validate these organ-chips as preclinical models.”
Organ chips: modeling complexity
At the forefront of organ-on-a-chip models are those used in toxicity testing, where the model replicates a healthy liver or kidney.3 “There's been some really good research beginning to validate those liver models which shows that, when liver or kidney models are incorporated into a preclinical testing pipeline, they actually save money for pharma because they have better predictive power than animals,” says Knight. “But increasingly, organ chips are being explored for their potential in efficacy testing, but we don't yet have enough good, validated models for all the different conditions that are usable by industry.”
There are now a wide range of commercial OOAC platforms available – these include systems with the ability to apply mechanical or physiological forces – which can then be bioengineered to reproduce the characteristics of a model or disease, but this requires building in a level of complexity that is often technologically challenging.
In the Molecular Cell Biomechanics Lab at the University of California Berkeley, Dr. Amin Valiei is taking a step-by-step approach to reconstructing one of the most complex human systems on a chip – the human gut microbiome.
“Most research has focused on finding correlations between microbiome composition and diseases in people using sequencing techniques of fecal samples, but what is lacking is a mechanistic understanding of how the microbial composition and structure contributes to these diseases,” said Valiei. “Our aim is to make a model of the intestine and of intestinal microbiology in the lab. By taking advantage of microfluidic technology, an OOAC will model transport processes such as hydrodynamics, chemical gradients and the continuous flow of nutrients that occurs in the gut, meaning we can address a lot of the shortcomings of simpler techniques such as plate cultures or organoids, which cannot replicate these physiological conditions.”
There are a lot of technological hurdles to creating a realistic gut microbiome OOAC, but improved gut-on-a-chip models now allow prolonged coculture of microbiota and human cells,4 and increasing coverage of microbial community databases and advancements in sequencing techniques are providing a better picture of the gut’s microbial community. Now, Valiei says, they need to build on this to understand the microbe types and culture conditions that reproduce what is seen in humans.
“For example, one aspect is that host cells generally secrete a mucus layer that keeps the bacteria at a distance within the gut, but this secretion is commonly lower in model cells compared to the real human cells,” explained Valiei. “So, we are focusing on modeling microbial structures inside the intestine, understanding and reproducing the optimal conditions to keep these microbes alive. We also aim to induce microbes to form microbial communities in co-cultures with human cells and use computational modeling to understand host-microbe and microbe-microbe interactions.”
The goal is to build a stable microbial community that has a composition similar to gut communities and use this knowledge first to better understand disease but also later as a tool for precision medicine, where different people’s fecal samples are tested and the signature of the microbial community can be used to predict, prevent and potentially treat disease.
At the Centre for Predictive in vitro Models at Queen Mary University London, Knight and colleagues are also focused on a range of highly complex OOACs, including a breast cancer bone metastasis model. “You’ve got bone which is a complex tissue and is mechanosensitive, three different bone cell types, vascularization, and then you have to add on the tumor and the tumor microenvironment, so that's a pretty complicated model to build,” said Knight. “We think we're getting closer to doing that.”
Another complicated example is their model of a human synovium-on-a-chip recently reported for studying inflammation in arthritis.5 This chip comprises synovial cells, blood vessels and circulating immune cells to allow researchers to study how immune cells migrate into the joint, causing inflammation. “All the models we're producing allow both fundamental research into disease processes and screening of new compounds and therapeutics,” explained Knight. So, we've produced the vascularized synovium-on-a-chip model, but we're also producing models of the synovial joint incorporating the important interactions between tissues such as cartilage, bone and synovium. These complex in vitro models enable researchers to build up the complexity depending on the question for which the organ-chip is to be used.”
This complexity is not only limited to building in biological features, but also building in diversity. “With medicines currently tested in clinical trials in humans, it's difficult to capture the full diversity of our human population. And it's not just genetic diversity, it’s our lifestyle exposures that also makes us different,” said Knight. “So, we also need to work out how to build that inherent diversity into our OOAC models.”
Validating OOACs as efficacy models
The next major challenge in realizing the potential of OOACs in preclinical development is their validation, i.e., ensuring they are non-inferior to conventional animal models. This is directly related to their complexity, says Knight. “How much complexity do we need to replicate in an organ chip, for it to be effective? That's a difficult question to answer. You can't replicate everything. It would be big, hugely expensive and take ages to develop. But I think everybody agrees we need more complexity to this than a monolayer of one cell type grown on a flat sheet of plastic.”
The first step is for stakeholders – including academia, industry and regulators – to come together to work out how to validate these models for the different organs and different diseases and test whether these OOAC models are better at predicting whether a drug is going to work compared to the gold standard animal model.
The Queen Mary Centre for Predictive in vitro Models brings together academics with representatives from over 50 companies working in this field. This includes major biopharma industry, key regulators such as the Medicines and Healthcare Regulatory Agency, and industry developing next-generation organ-chip technology. By working in partnership, the group is trying to address these questions to ensure that OOAC technology is going to be robust enough to test whether new therapies will be safe and effective in patients.
“What we need is a consistent approach for validating the models, so you know you've got an approved model that accurately replicates disease biology and can be used for preclinical testing. These models have huge potential to support therapeutic innovation and not just for small-molecule drugs, but also gene therapy, stem cell therapy and all sorts of emerging new health solutions,” said Knight. “If we could use OOACs to better predict which of these solutions will be successful in clinical trials, and even predict which treatment is best for an individual patient, then we'd drive better healthcare and save time and money. As a result it becomes economically viable to develop other therapeutics, even for prevalent diseases. Ultimately, you'd get better and better delivery of new therapeutics for the benefit of us all.”
About the interviewees:
Martin Knight is a professor of mechanobiology and a bioengineer based at Queen Mary University of London. His research examines how living cells and tissues within our bodies sense and respond to mechanical forces with particular focus on organ-chip technology.
Amin Valiei is a postdoctoral fellow at the University of California, Berkeley. His research focuses on the development of advanced in vitro and in silico models to gain insights into the conditions that promote the health of the human gut microbiome and explore potential causes and treatments for imbalances occurring within this complex system.